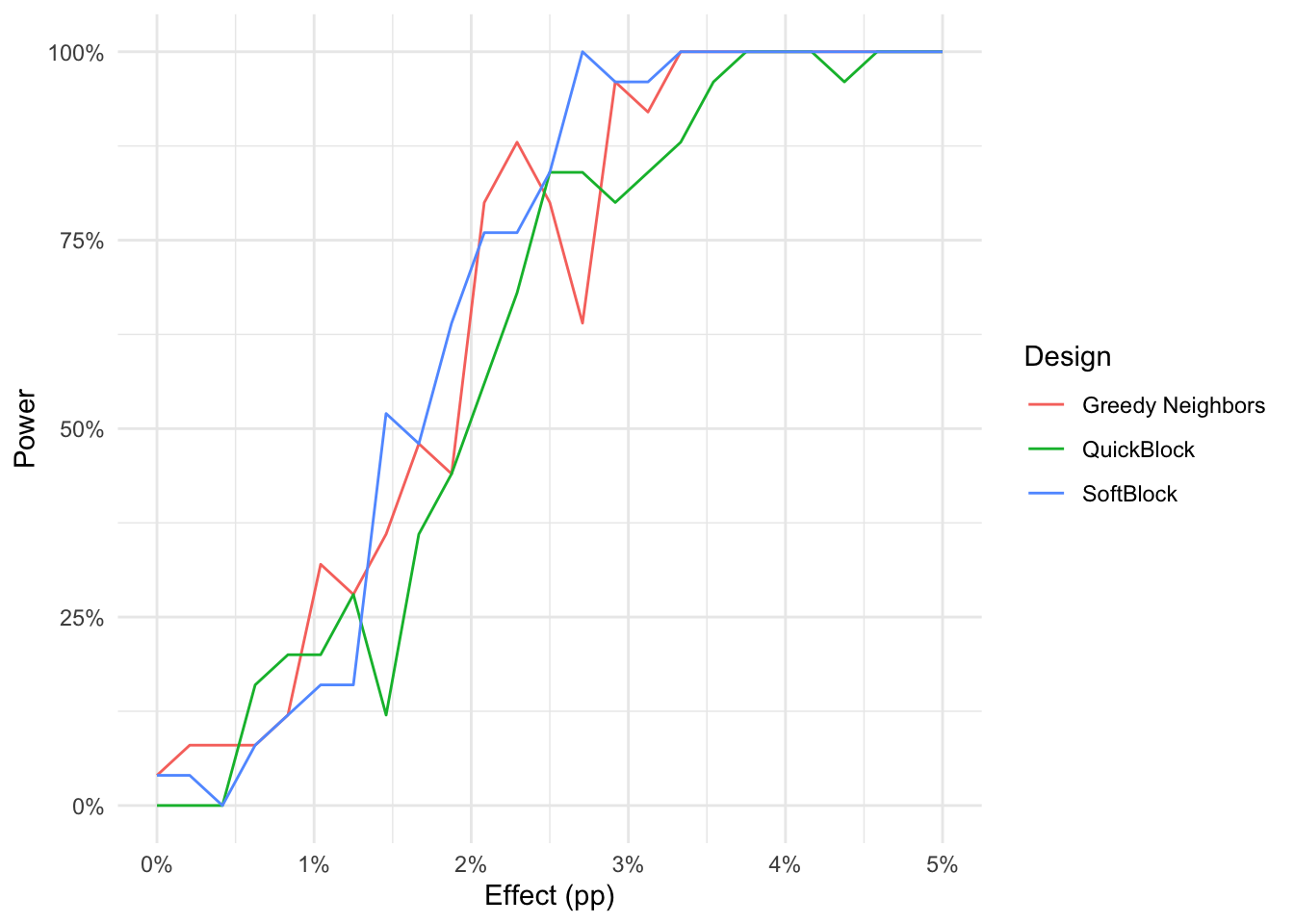
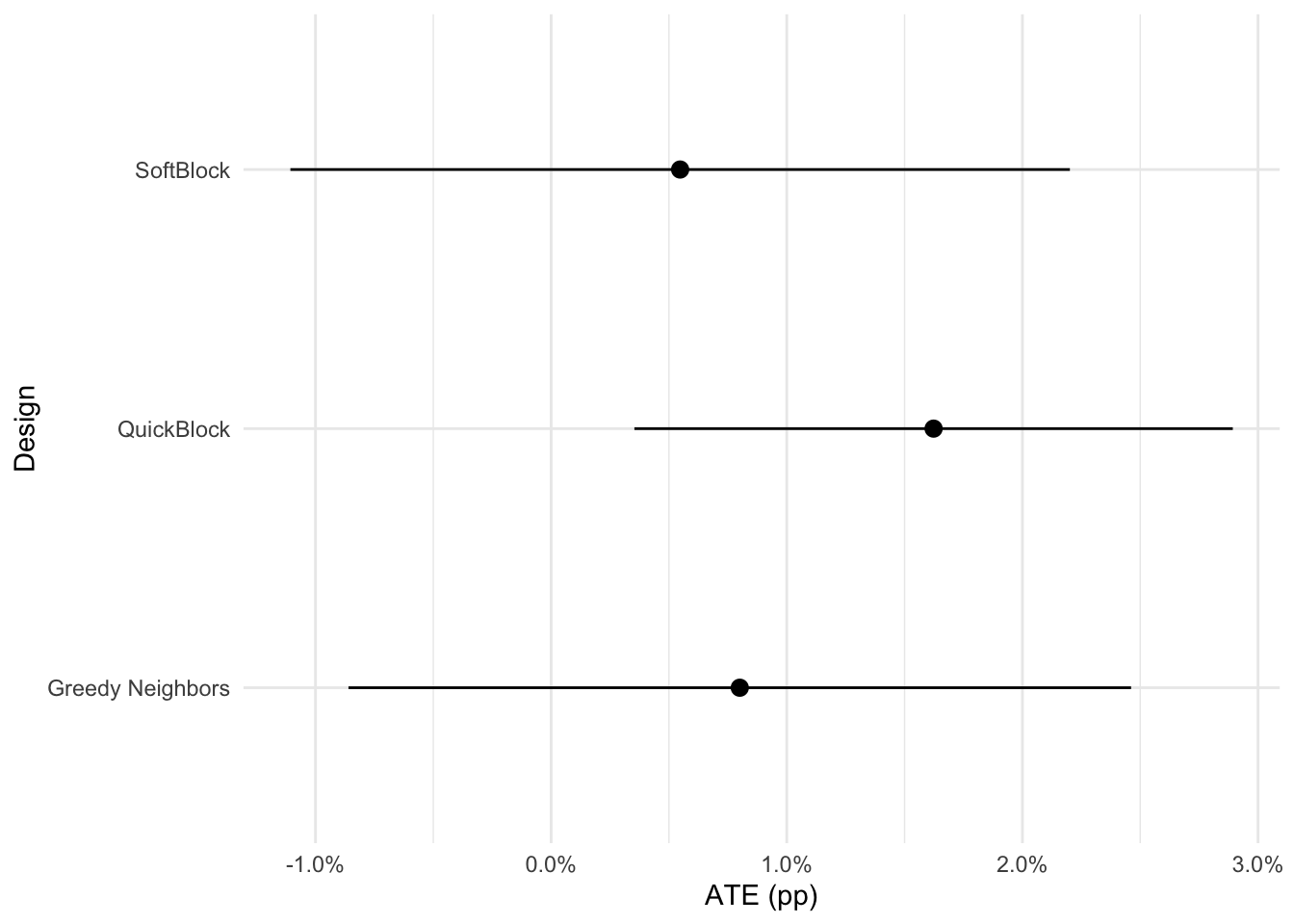
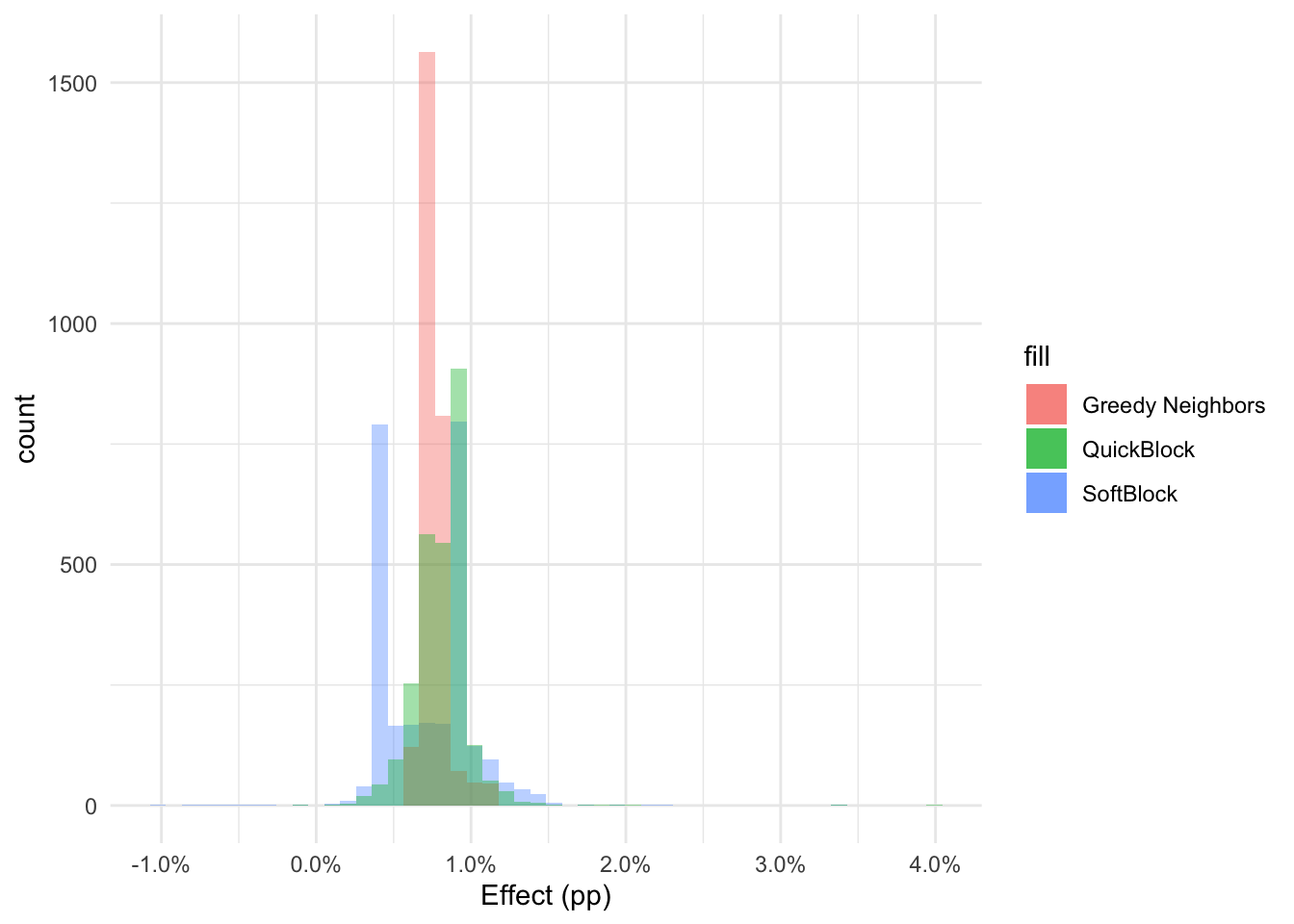
library(Matrix)
library(igraph)
library(FNN)
library(hash)
assign_greedily <- function(graph) {
adj_mat = igraph::as_adjacency_matrix(graph, type="both", sparse=TRUE) != 0
N = nrow(adj_mat)
root_id = make.keys(sample(N, 1))
a = rbinom(1, 1, 0.5)
visited = hash()
random_order = make.keys(sample(N))
unvisited = hash(random_order, random_order)
colors = hash()
stack = hash()
stack[[root_id]] <- a
tentative_color = rbinom(N, 1, 0.5)
while ((!is.empty(unvisited)) || (!is.empty(stack))) {
if (is.empty(stack)) {
cur_node = keys(unvisited)[1]
del(cur_node, unvisited)
color = tentative_color[as.integer(cur_node)]
} else {
cur_node = keys(stack)[1]
color = stack[[cur_node]]
del(cur_node, stack)
del(cur_node, unvisited)
}
visited[[cur_node]] = cur_node
colors[[cur_node]] = color
children = make.keys(which(adj_mat[as.integer(cur_node), ]))
for (child in children) {
if(has.key(child, unvisited)) {
stack[[child]] = 1 - color
}
}
}
values(colors, keys=1:N)
}
assign_softblock <- function(.data, cols, .s2=2, .neighbors=6) {
expr <- rlang::enquo(cols)
pos <- tidyselect::eval_select(expr, data = .data)
df_cov <- rlang::set_names(.data[pos], names(pos))
cov_mat = scale(model.matrix(~.+0, df_cov))
N = nrow(cov_mat)
st = lubridate::now()
knn = FNN::get.knn(cov_mat, k=.neighbors)
st = lubridate::now()
knn.adj = Matrix::sparseMatrix(i=rep(1:N, .neighbors), j=c(knn$nn.index), x=exp(-c(knn$nn.dist) / .s2))
knn.graph <- graph_from_adjacency_matrix(knn.adj, mode="plus", weighted=TRUE, diag=FALSE)
E(knn.graph)$weight <- (-1 * E(knn.graph)$weight)
st = lubridate::now()
mst.graph = igraph::mst(knn.graph)
E(mst.graph)$weight <- (-1 * E(mst.graph)$weight)
st = lubridate::now()
assignments <- assign_greedily(mst.graph)
.data$treatment <- assignments
attr(.data, "laplacian") <- igraph::laplacian_matrix(mst.graph, normalize=TRUE, sparse=TRUE)
.data
}
assign_greedy_neighbors <- function(.data, cols) {
expr <- rlang::enquo(cols)
pos <- tidyselect::eval_select(expr, data = .data)
df_cov <- rlang::set_names(.data[pos], names(pos))
cov_mat = scale(model.matrix(~.+0, df_cov))
N = nrow(cov_mat)
knn = FNN::get.knn(cov_mat, k=1)
knn.adj = Matrix::sparseMatrix(i=1:N, j=c(knn$nn.index), x=c(knn$nn.dist))
knn.graph <- graph_from_adjacency_matrix(knn.adj, mode="plus", weighted=TRUE, diag=FALSE)
assignments <- assign_greedily(knn.graph)
.data$treatment <- assignments
attr(.data, "laplacian") <- igraph::laplacian_matrix(knn.graph, normalize=TRUE, sparse=TRUE)
.data
}
assign_matched_pairs <- function(.data, cols, .s2=2, .neighbors=6) {
expr <- rlang::enquo(cols)
pos <- tidyselect::eval_select(expr, data = .data)
df_cov <- rlang::set_names(.data[pos], names(pos))
cov_mat = scale(model.matrix(~.+0, df_cov))
N = nrow(cov_mat)
knn = FNN::get.knn(cov_mat, k=.neighbors)
knn.adj = Matrix::sparseMatrix(i=rep(1:N, .neighbors), j=c(knn$nn.index), x=exp(-c(knn$nn.dist) / .s2))
knn.graph <- graph_from_adjacency_matrix(knn.adj, mode="plus", weighted=TRUE, diag=FALSE)
E(knn.graph)$weight <- (-1 * E(knn.graph)$weight)
mwm.graph = igraph::max_bipartite_match(knn.graph)
E(mwm.graph)$weight <- (-1 * E(mwm.graph)$weight)
assignments <- assign_greedily(mwm.graph)
.data$treatment <- assignments
attr(.data, "laplacian") <- igraph::laplacian_matrix(mwm.graph, normalize=TRUE, sparse=TRUE)
.data
}
# library(tibble)
# data = tibble(
# x1=runif(10),
# x2=runif(10),
# x3=rbinom(10, 1, 0.5)
# )
# library(dplyr)
# library(tidyr)
# library(ggplot2)
# data %>% assign_softblock(c(x1, x2)) -> newdata
# ggplot(newdata, aes(x=x1, y=x2, color=factor(treatment), shape=factor(x3))) + geom_point() + theme_minimal()
# newdata %>%
# attr("laplacian") %>%
# ifelse(lower.tri(.), ., 0) %>%
# as_tibble() -> adj_df
# names(adj_df) <- paste0(1:ncol(adj_df))
# adj_df %>%
# group_by(id_1=as.character(row_number())) %>%
# gather(id_2, weight, -id_1) %>%
# filter(weight != 0) %>%
# mutate(id_2=as.character(id_2), id=paste(id_1, id_2, sep='-')) -> adj_df
# locs = newdata %>% mutate(id=as.character(row_number())) %>% select(id, x1, x2, x3)
# edges=bind_rows(
# adj_df %>% inner_join(locs, by=c('id_1'='id')),
# adj_df %>% inner_join(locs, by=c('id_2'='id'))
# ) %>% arrange(id) %>% ungroup()
# pp = ggplot(newdata, aes(x=x1, y=x2)) +
# geom_line(aes(group=id, size=1), data=edges, color='grey') +
# geom_point(aes(color=factor(treatment), shape=factor(x3), size=2), alpha=.9) +
# scale_size_continuous(range=c(1, 3)) +
# theme_minimal() + theme(legend.position='none')
# print(pp)